Establishment of a collection of commensal bacterial strains of chicken for exploration as potential probiotic candidates
Case study
Establishment of a collection of commensal bacterial strains of chicken for exploration as potential probiotic candidates

Boehringer-Ingelheim Animal Health develops products and solutions to improve animal health, including in the field of poultry production.
The chicken industrial production faces strong challenges to maintain quality and productivity standards.
In this context, the chicken gastrointestinal microbiota has emerged as an important parameter that should be taken into account to increase animals’ performances. It can potentially improve nutrition efficacy as well as resistance to infections or colonization by zoonotic infectious agents.
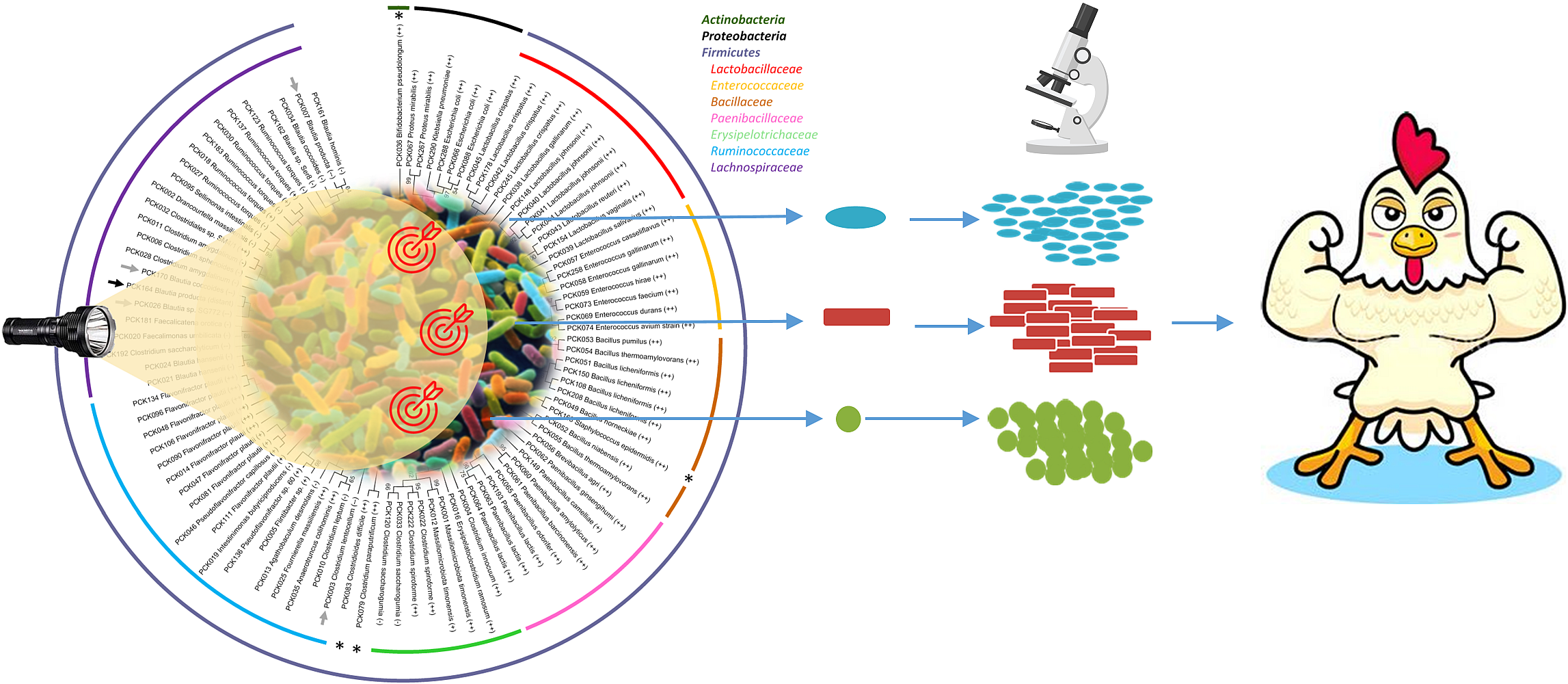
16S rRNA gene repertoire analysis was used to investigate the caecal microbiota taken from 7-day-old chicks from two conventional high performance farms but also from a free-range farm presenting lower performance indicators.
In parallel, a wide variety of commensal bacteria species were cultured from chicken samples from the high performance farms. Once identified and biobanked, these strains were explored for several important characteristics including bacteriocin production, short chain fatty acid production, capacity to form spores, biofilm formation, swimming and swarming, amyloids production, resistance to bile acids…
Significant differences were found between the caecal microbiota compositions of chickens from high vs low performance farms, including the presence of the potentially pathogenic species Campylobacter jejuni in the latter.
A collection of 350 isolates corresponding to at least 64 different species was established, with a significant proportion of potentially new species in the collection.
Based on their capacity to inhibit C. jejuni growth as well as other important traits, forty isolates were selected for full genome sequencing. Additional bioinformatic analyses were performed to facilitate the design of several bacterial consortia that were evaluated in in vivo assays.
The non-targeted culturomics approach developed in this collaborative study can be used to build collections of commensal bacteria from any livestock animal. It can also be adapted to cultivate bacteria from other kinds of samples such as insects, environmental samples etc. The rapid phenotypic characterization of the isolated strains can be based on the tests already developed but other tests aiming, for example, at detecting specific activities and/or the production of molecules of interest can also be set up.