Comparative analysis of supervised integrative methods for multi-omics data
Case study
Comparative analysis of supervised integrative methods for multi-omics data

Recent advances in sequencing, mass spectrometry and cytometry technologies have enabled researchers to collect multiple omics data from a single sample. These large datasets have led to a growing consensus that a holistic approach was needed to identify new candidate biomarkers and unveil mechanisms underlying disease aetiology, key to precision medicine. In collaboration with a US partner, this project aimed at landscaping and benchmarking supervised integrative approaches.
Due to the relative novelty of the field, numerous challenges remain in multi-omics analysis among which:
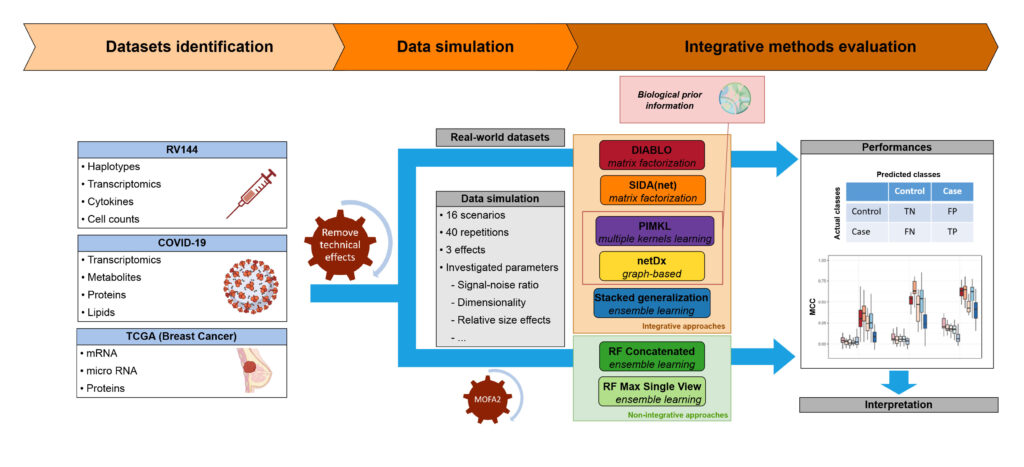
BIOASTER reviewed and selected cutting-edge machine learning methods,representative of the main families of integrative approaches (matrix factorization,multiple kernel methods, ensemble learning and graph-based approaches). Methods were subsequently evaluated on both simulated and real world datasets; the latter were carefully selected to cover various medical applications (infectious diseases, oncology,and vaccine).
Integrative approaches showed comparable or higher performances on simulations and outperformed non-integrative methods on real-world data. More specifically, multiple kernel and matrix factorization demonstrated a strong ability to uncover modest effectsin high dimensionality settings.
The expertise acquired in this project will help BIOASTER and its partners refine both biomarker discovery and decipher molecular function underlying new mode of actions.
After a first study on unsupervised integrative approaches (ref 2019), we are thrilled to announce the 2024 publication of our article in “Briefings in Bioinformatics” (ref 2024) in which we investigate the performances of six popular supervised integrative methods in three medical applications. This study marks a significant milestone for our team.
Article: https://doi.org/10.1093/bib/bbae331